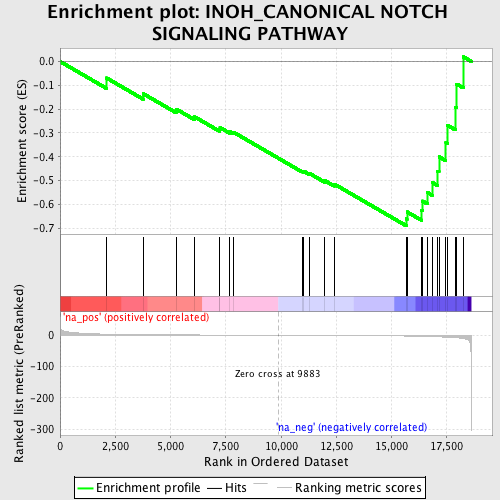
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_DMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | INOH_CANONICAL NOTCH SIGNALING PATHWAY |
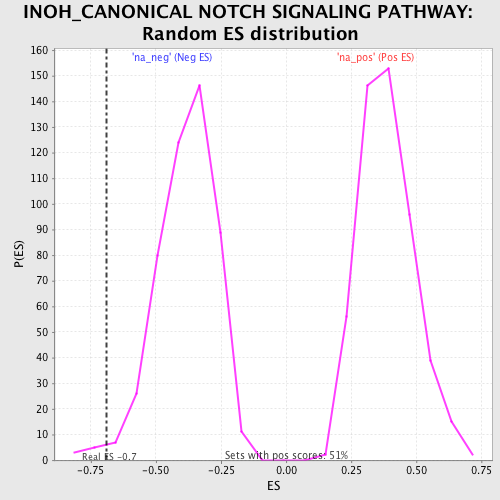
| Enrichment Score (ES) | -0.6904368 |
| Normalized Enrichment Score (NES) | -1.7885667 |
| Nominal p-value | 0.01629328 |
| FDR q-value | 0.28727135 |
| FWER p-Value | 0.996 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MFNG | 2102 | 3.495 | -0.0691 | No | ||
| 2 | JAG2 | 3759 | 1.852 | -0.1349 | No | ||
| 3 | NOTCH4 | 5254 | 1.240 | -0.1997 | No | ||
| 4 | NOTCH2 | 6092 | 0.977 | -0.2324 | No | ||
| 5 | DLL3 | 7227 | 0.664 | -0.2851 | No | ||
| 6 | LFNG | 7235 | 0.663 | -0.2771 | No | ||
| 7 | RFNG | 7677 | 0.545 | -0.2940 | No | ||
| 8 | FBXW7 | 7836 | 0.500 | -0.2962 | No | ||
| 9 | NOTCH3 | 10955 | -0.276 | -0.4604 | No | ||
| 10 | JAG1 | 11026 | -0.296 | -0.4605 | No | ||
| 11 | ADAM10 | 11302 | -0.374 | -0.4706 | No | ||
| 12 | DTX3 | 11980 | -0.554 | -0.5000 | No | ||
| 13 | DLL4 | 12405 | -0.682 | -0.5143 | No | ||
| 14 | DLL1 | 15681 | -2.380 | -0.6605 | Yes | ||
| 15 | RBPJ | 15704 | -2.406 | -0.6314 | Yes | ||
| 16 | ADAM17 | 16353 | -3.263 | -0.6253 | Yes | ||
| 17 | DTX1 | 16396 | -3.336 | -0.5856 | Yes | ||
| 18 | NUMB | 16624 | -3.805 | -0.5500 | Yes | ||
| 19 | FURIN | 16841 | -4.282 | -0.5078 | Yes | ||
| 20 | PSEN1 | 17090 | -4.855 | -0.4601 | Yes | ||
| 21 | DTX2 | 17151 | -5.033 | -0.4000 | Yes | ||
| 22 | NOTCH1 | 17461 | -5.950 | -0.3418 | Yes | ||
| 23 | CREBBP | 17538 | -6.187 | -0.2681 | Yes | ||
| 24 | PSEN2 | 17891 | -7.601 | -0.1915 | Yes | ||
| 25 | MAML1 | 17950 | -7.943 | -0.0948 | Yes | ||
| 26 | ITCH | 18253 | -10.384 | 0.0195 | Yes |